

 |
 |
 |
 Tractography of the major white matter fibers is an essential tool for understanding similarities and differces between brains. Tractography results
can inform us about any particular brain or be compared directly, for instance using
connectivity blueprint matching.
Tractography of the major white matter fibers is an essential tool for understanding similarities and differces between brains. Tractography results
can inform us about any particular brain or be compared directly, for instance using
connectivity blueprint matching.
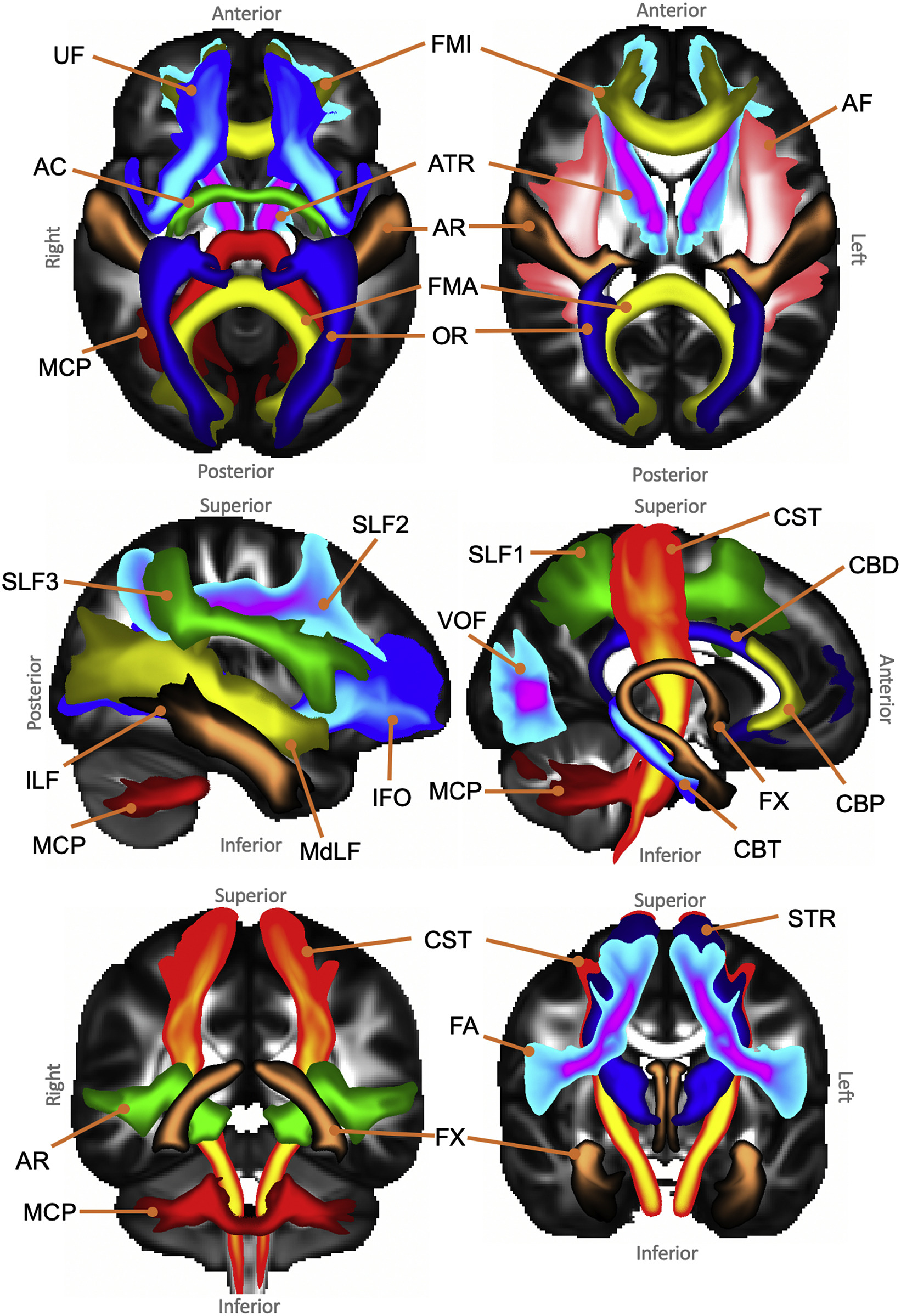
To standize the way particular tracts are identified we use FSL's Xtract, formerly known as autoPtx. This is a simple tool that takes predefined mask images in a standard space and runs probabilistic tractography in native/subject space. An initial autoPtx library for the human brain was defined by Marius de Groot, but we have been working on new, expanded libraries together with Stam Sotiropoulos and Saad Jbabdi.
We have built comparable libraries for the human brain and various other primates, using similar protocols to aid comparison between species. This page presents the different libraries as they have been used in our papers and contains links to the relevant data repositories in reverse chronological order. Since our understanding of these brains continues to evolve, the libraries are updated regularly. The same tract that be defined differently in different libraries, as our knowledge expands. In effect, the Xtract libraries can be seen as a tractography spreadsheet: if you don't like a particular definition we have, you can simply supply your own and see how the results change.
If you have feedback on our libraries or have developed your own, please contact us.
Core Xtract paper
Following our initial human/macaque paper, the core paper describing the Xtract tool by Shaun Warrington et al. validates an extended library using the Human Connectome Project and UK Biobank data; their projections and laterality generally match those described in the literature.
The Xtract paper is published as Warrington et al., 2020, NeuroImage and the recipes are available on the WIN GitLab
Human/macaque connectivity blueprint matching
Our first autoPtx library defined 39 tracts in the human and macaque brain. These tracts were used to create a common space between the two species that allowed us to match cortical vertices based on their connectivity fingerprint with the tracts. We called this technique 'connectivity blueprint matching'. It was first described in our connectivity blueprint matching paper.
The autoPtx protocols used in this study are part of the data sharing collection.
Chimpanzee (Pan troglodytes)
Recipes for the chimpanzee were developed by Katherine Bryant in Yerkes29 ("Chimplate") space. Recipes for 45 tracts were created, along with comparable recipes for the human and macaque to allow direct comparison.
The recipes and their tracts are presented in Bryant et al., 2020, bioRxiv, following an earlier exploratory analysis in Mars et al., 2019, Brain Struct Funct.
The protocols used in this study are part of the data sharing collection.